Hi,
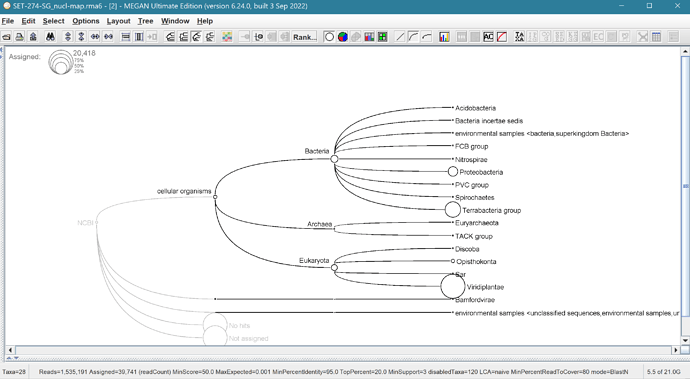
I’m having an issue with the MEGAN6 - Ultimate Edition trial. I want to look at the KEGG data for some metagenomic libraries, but I can’t get any taxa to assign when using the functional database mapping file. When using the regular taxonomic file (megan-nucl-Jan201.db is what I am currently using) importing data from BLAST, reads assign normally as I’ve done for many libraries.
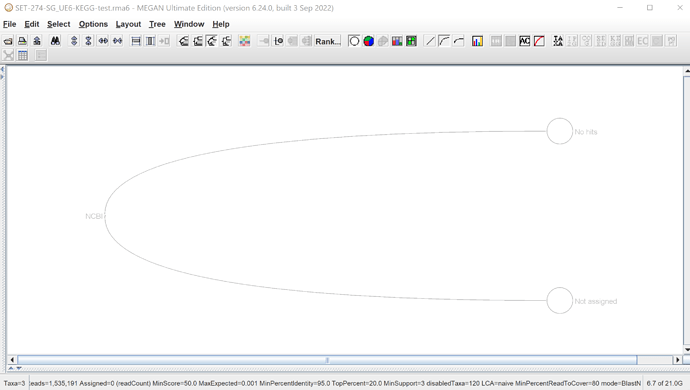
However, when I try to use the Ultimate Edition specific mapping file for functional analysis (megan-map-Feb2022-ue.db), using identical LCA parameters, nothing assigns.
Any suggestions for how to fix this? I’m trying to run the ue mapping file with extended mode to see if that fixes it, but it takes many more hours to run that way instead of in fast mode when using much larger files (with more blast assigned reads). So if there’s another solution, that would be really helpful. I assume it has to do with the formatting of the BLAST accessions, but I’m not sure how to fix it. I’ve generated my BLAST files with default output parameters.
Update: extended mode didn’t work either. Nothing was assigned.
blastn -query $f -out “$b”.MegaBLAST.50hits.113-2019.fullout.blast -db /scratch/blastdb/nt -num_alignments 50 -max_hsps 1 -num_threads 5 -evalue 0.001
I’ve uploaded the example fasta/blast file here if it’s helpful to see what’s going on: https://drive.google.com/drive/folders/1Yu_uMWOEikZPZJRw6YGzUyZ8xYF_lT2a?usp=sharing
Thanks for your help!