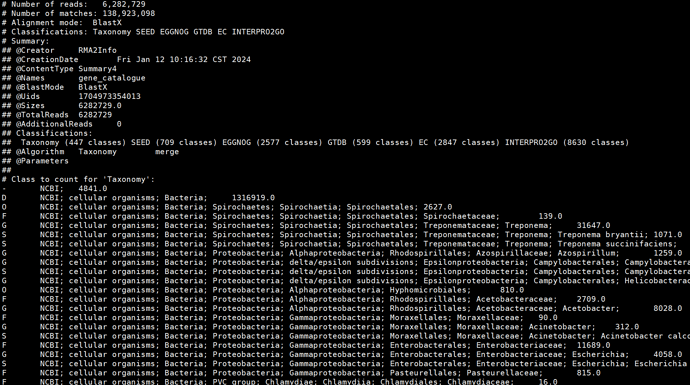
Hello everyone, I got the species annotation file through diamond and megan6, but it only has about 400 species annotation information. What’s going on?
Uploading: image.png…
This is the command I ran:
diamond blastx -d ./diamond_index/Bacteria_nr.dmnd -q ./UniqGenes/Unigenes.CDS.cdhit.fa -o contigs.daa -f 100
daa-meganizer -i ./MEGAN6/contigs.daa -mdb ./MEGAN6/megan-map-Feb2022.db
/megan/tools/daa2info -i ./MEGAN6/contigs.daa -c2c Taxonomy -r2c Taxonomy --paths true --list ture -r true -mro true > ./MEGAN-taxonomy3.txt
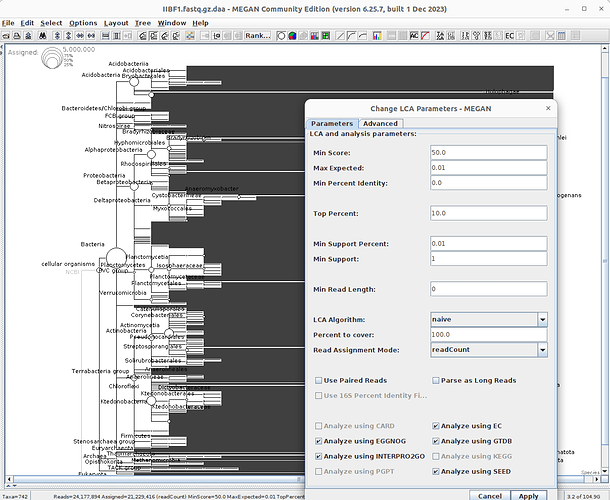
Hi, may be you can try to modify some LCA parameters like the minimun supported percent to a small number (by default is 0.01). Remember that MEGAN6 by default perform some filters to assessment the best taxonomy classification.
Hi good bro, I’m just a beginner, how should I change my command?
i know that its possible do this action using command line but you can try using GUI application of MEGAN.T
ry to do this in the options section
Oh, I see, so what parameters can I try to change? Allows me to obtain more species annotation information
My recomendation its try to put a small value of minimun supported percent. If you want for example to see all assignaments without this filter, you can try put this value to 0, but remember that doing this can increase the number of false assignments. you have to decide what kind of value you want to put based on the filter keeps by default all the reads that contain at least 1% of assignments (default is 0.01)