I created a multi-fasta genome reference (of parasitic-worms). The header does not state any taxonomic classification.
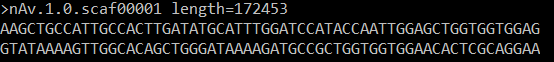
The MALT-build output looked fine.
When I run a malt build + screen my samples this is the megan output:
![]()
My best guess is, that there is a step I did not take. I would have thought that the reference index with the megan-nucl-Feb2022.db.zip would be able to sort the taxonomy and the header of the concatenated genomic sequences are not important.
I anyone could give me a push in the right direction? I would be very grateful.