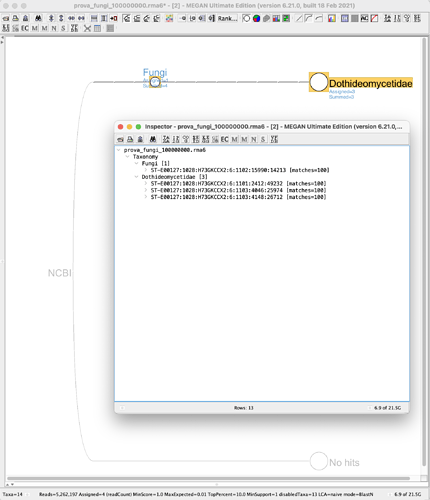
BLASTN 2.11.0+
Reference: Zheng Zhang, Scott Schwartz, Lukas Wagner, and Webb
Miller (2000), “A greedy algorithm for aligning DNA sequences”, J
Comput Biol 2000; 7(1-2):203-14.
Database: Internal transcribed spacer region (ITS) from Fungi type and
reference material
12,165 sequences; 7,153,958 total letters
Query= ST-E00127:1028:H73GKCCX2:6:1101:3153:1854 1:N:0:CAGCTA
Length=151
***** No hits found *****
Lambda K H
1.33 0.621 1.12
Gapped
Lambda K H
1.28 0.460 0.850
Effective search space used: 896804090
Query= ST-E00127:1028:H73GKCCX2:6:1101:3234:1854 1:N:0:CAGCTA
Length=151
***** No hits found *****
Lambda K H
1.33 0.621 1.12
Gapped
Lambda K H
1.28 0.460 0.850
Effective search space used: 896804090
Query= ST-E00127:1028:H73GKCCX2:6:1101:4148:1854 1:N:0:CAGCTA
Length=151
***** No hits found *****
Lambda K H
1.33 0.621 1.12
Gapped
Lambda K H
1.28 0.460 0.850
Effective search space used: 896804090
Query= ST-E00127:1028:H73GKCCX2:6:1101:4229:1854 1:N:0:CAGCTA
Length=151
***** No hits found *****
Lambda K H
1.33 0.621 1.12
Gapped
Lambda K H
1.28 0.460 0.850
Effective search space used: 896804090
#AFTER A LOT OF THIS “NO HITS” (I have searched the hits through sed command)
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 287 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 346
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 347 TCAAGCCTCGCT 358
NR_163324.1 Cladosporium entadae MFLUCC 17-0919 ITS region; from TYPE material
Length=488
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 269 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 328
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 329 TCAAGCCTCGCT 340
NR_158360.1 Cladosporium endophytica MFLUCC 17-0599 ITS region; from TYPE
material
Length=581
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 317 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 376
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 377 TCAAGCCTCGCT 388
NR_156582.1 Ramularia mali CBS 129581 ITS region; from TYPE material
Length=588
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 347 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 406
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 407 TCAAGCCTCGCT 418
NR_156581.1 Ramularia glennii CBS 129441 ITS region; from TYPE material
Length=588
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 347 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 406
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 407 TCAAGCCTCGCT 418
NR_156348.1 Cladosporium domesticum CPC 22307 ITS region; from TYPE material
Length=555
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 302 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 361
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 362 TCAAGCCTCGCT 373
NR_156347.1 Cladosporium coloradense CPC 22238 ITS region; from TYPE material
Length=625
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 377 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 436
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 437 TCAAGCCTCGCT 448
NR_154953.1 Ramularia vallisumbrosae CBS 272.38 ITS region; from TYPE material
Length=593
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 350 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 409
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 410 TCAAGCCTCGCT 421
NR_154951.1 Ramularia rubella CPC 25911 ITS region; from TYPE material
Length=569
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 347 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 406
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 407 TCAAGCCTCGCT 418
NR_154950.1 Ramularia osterici CPC 10750 ITS region; from TYPE material
Length=578
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 349 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 408
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 409 TCAAGCCTCGCT 420
NR_154949.1 Ramularia lamii var. lamii CBS 108970 ITS region; from TYPE material
Length=588
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 347 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 406
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 407 TCAAGCCTCGCT 418
NR_154948.1 Ramularia heraclei CBS 108969 ITS region; from TYPE material
Length=588
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 347 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 406
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 407 TCAAGCCTCGCT 418
NR_154944.1 Ramularia collo-cygni CBS 101180 ITS region; from TYPE material
Length=588
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 349 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 408
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 409 TCAAGCCTCGCT 420
NR_154943.1 Ramularia armoraciae CBS 241.90 ITS region; from TYPE material
Length=588
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 347 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 406
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 407 TCAAGCCTCGCT 418
NR_154942.1 Ramularia abscondita ITS region; from TYPE material
Length=588
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 347 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 406
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 407 TCAAGCCTCGCT 418
NR_154919.1 Ramularia hydrangeicola KACC 43597 ITS region; from TYPE material
Length=495
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 259 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 318
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 319 TCAAGCCTCGCT 330
NR_154917.1 Ramularia pusilla CBS 124973 ITS region; from TYPE material
Length=591
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 349 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 408
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 409 TCAAGCCTCGCT 420
NR_154911.1 Ramularia haroldporteri CPC 16296 ITS region; from TYPE material
Length=598
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 356 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 415
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 416 TCAAGCCTCGCT 427
NR_154901.1 Ramularia cynarae CPC 18426 ITS region; from TYPE material
Length=593
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 371 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 430
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 431 TCAAGCCTCGCT 442
NR_154899.1 Ramularia stellariicola ITS region; from TYPE material
Length=645
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 350 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 409
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 410 TCAAGCCTCGCT 421
NR_154898.1 Ramularia acroptili CBS 120252 ITS region; from TYPE material
Length=645
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 350 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 409
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 410 TCAAGCCTCGCT 421
NR_152307.1 Cladosporium chasmanthicola CPC 21300 ITS region; from TYPE material
Length=589
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 360 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 419
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 420 TCAAGCCTCGCT 431
NR_152296.1 Cladosporium pseudochalastosporoides ITS region; from TYPE material
Length=599
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 360 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 419
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 420 TCAAGCCTCGCT 431
NR_152295.1 Cladosporium penidielloides ITS region; from TYPE material
Length=599
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 365 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 424
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 425 TCAAGCCTCGCT 436
NR_152286.1 Cladosporium angustiherbarum ITS region; from TYPE material
Length=598
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 359 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 418
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 419 TCAAGCCTCGCT 430
NR_152308.1 Cladosporium welwitschiicola CPC 18648 ITS region; from TYPE
material
Length=535
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 300 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 359
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 360 TCAAGCCTCGCT 371
NR_145121.1 Ramularia eucalypti ITS region; from TYPE material
Length=588
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 348 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 407
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 408 TCAAGCCTCGCT 419
NR_145097.1 Ramularia proteae CBS 112161 ITS region; from TYPE material
Length=683
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 390 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 449
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 450 TCAAGCCTCGCT 461
NR_145125.1 Ramularia hydrangeae-macrophyllae CBS 122273 ITS region; from
TYPE material
Length=612
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 371 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 430
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 431 TCAAGCCTCGCT 442
NR_154947.1 Ramularia geranii CBS 160.24 ITS region; from TYPE material
Length=612
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 370 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 429
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 430 TCAAGCCTCGCT 441
NR_154946.1 Ramularia euonymicola CBS 113308 ITS region; from TYPE material
Length=480
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 258 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 317
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 318 TCAAGCCTCGCT 329
NR_154945.1 Ramularia digitalis-ambiguae CBS 297.37 ITS region; from TYPE
material
Length=592
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 368 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 427
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 428 TCAAGCCTCGCT 439
NR_154941.1 Ramularia citricola CPC 26192 ITS region; from TYPE material
Length=531
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 289 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 348
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 349 TCAAGCCTCGCT 360
NR_152300.1 Cladosporium aggregatocicatricatum ITS region; from TYPE material
Length=641
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 402 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 461
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 462 TCAAGCCTCGCT 473
NR_152299.1 Cladosporium rhusicola ITS region; from TYPE material
Length=639
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 400 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 459
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 460 TCAAGCCTCGCT 471
NR_152298.1 Cladosporium puyae CBS 274.80A ITS region; from TYPE material
Length=638
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 399 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 458
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 459 TCAAGCCTCGCT 470
NR_152294.1 Cladosporium aciculare ITS region; from TYPE material
Length=635
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 396 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 455
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 456 TCAAGCCTCGCT 467
NR_152293.1 Cladosporium parapenidielloides ITS region; from TYPE material
Length=590
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 360 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 419
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 420 TCAAGCCTCGCT 431
NR_152297.1 Cladosporium versiforme ITS region; from TYPE material
Length=536
Score = 134 bits (72), Expect = 4e-32
Identities = 72/72 (100%), Gaps = 0/72 (0%)
Strand=Plus/Plus
Query 1 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 297 ACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCAC 356
Query 61 TCAAGCCTCGCT 72
||||||||||||
Sbjct 357 TCAAGCCTCGCT 368
NR_152291.1 Cladosporium longicatenatum ITS region; from TYPE material
Length=635
AND SO ON…