Hi,
I am new to megan and am trying to process blastn -task megablast results of 43 draft quality contigs ranging in size from 369 bases to 235 kilobases with the blast2lca tool. In the output, contigs <45k were assigned to a taxonomy by the algorithm while contigs >70k were not even though they had an abundance of hits. What might be the best way to get taxonomy assignments for these longer contigs? I am hesitant to use the long read algorithm since I don’t need or want a lot of open reading frame corrections and I don’t think it works with a blastn output… also, I’m not sure if this algorithm is in a stand alone tool. I was able to get two more contigs of size 73.6k and 99.8k assigned to taxonomy by filtering my blast results with -qcov_hsp_perc 1. I have tried other limiters such as -num_descriptions 100 (no effect) -num_alignments 50 (no effect) -max_hsps 5 (no effect) -culling_limit 1 (segmentation fault error). I also plan to try -word_size 40 and -evalue 0.00001 as well as tinkering with blast2lca options -ms, -me, and -top. But I’m flying blind since I don’t know why these longer contigs aren’t being assigned. Any help (both theoretical and practical) is appreciated 
Hello,
I wanted to follow up on this. blast2lca seems not to be detecting all the alignments in the blast file and often is only detecting a small fraction of them or none at all. I figured this out by installing the gui on my windows machine and watching the console. I did not test each contig individually, but I tested 3 of them: 1 that produced a taxonomy assignment and 2 that did not. Contig-22 has length-74,292 and has 22 alignments from 8 hits with between 1 and 4 alignments-per-hit. Megan is detecting -1 alignments and does not assign taxonomy. Contig-7 has length-103,590 and has 21 alignments from 9 hits with between 1 and 3 alignments-per-hit. Megan is detecting only 2 alignments and does not assign taxonomy. Contig-43 has length-106,343 and has 36 alignments from 8 hits with between 1 and 5 alignments-per-hit. Megan is detecting 2 alignments and does assign taxonomy. Am I hitting up against some max alignment length in the algorithm? Is there a way to tell which 2 alignments Megan is detecting in contigs 7 and 43? Additional alignments are not detected in long read mode. Please help!
Hello,
I was ultimately able to get taxonomic assignments for all 43 contigs by using the blast2lca tool packaged with version 5.13. I also attempted to install this earlier version on my windows machine to try and see what it was doing different but double clicking the installed executable doesn’t bring up a gui.
Please give me access to the data and I will look into this
Hello, I sent the blast result input files referenced in my second post to megan’AT’inf.uni-tuebingen.de on April 12, 2019. Did you receive them? Thanks so much for looking into this!
I just took a look at the files:
MEGAN can’t find the alignments for contig 22 because MEGAN is expecting the following line in the blast file:
Query=Contig_22_30.3655
The other two files that you sent me have that line present:
43.txt: Query= Contig_43_35.692
43.txt: Query= Contig_43_35.692
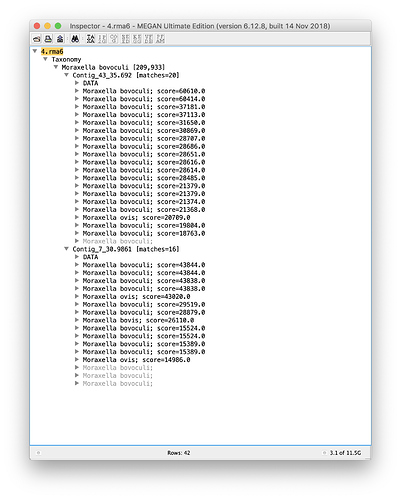
This is what I see after importing the three files into MEGAN:
So, a good number of alignments. Note that if you also provide the reads to MEGAN then you can also see how the reads align against the reference in a global view, in which you can also “turn on” the unaligned bases to see how much is not aligned.
Thanks so much for looking at them! Yes, it looks like I did not subdivide my multi-blast output correctly and omitted the string "Query= " in line 15 of “22.txt”. I’m sorry about that. Strangely, whereas previously the queries were assigned to species, they are now assigned only to order on my windows box. Clearly, I need to get to the bottom of multiple issues before I can provide a reproducible example (for Windows, at least). You mentioned the problem with 22.txt, but was contig 7 assigned to taxonomy for you? Here is my much more reproducible example for linux:
/home/aaron.dickey/megan/tools/blast2lca -i 3_contigs.txt -o 3_tax_assignments.txt
output of 3_tax_assignments.txt:
Contig_7_30.9861; ;
Contig_22_30.3655; ;
Contig_43_35.692; ;d__Bacteria; 100;p__Proteobacteria; 100;c__Gammaproteobacteria; 100;o__Pseudomonadales; 100;f__Moraxellaceae; 100;g__Moraxella; 100;s__Moraxella bovoculi; 100;
I will send the 3_contigs.txt & 3_tax_assignments.txt files to you. For the moment, I am continuing to use the blast2lca tool packaged with version 5.13. This produces taxonomy assignments for all 43 contigs. For 5.13, I use a tab separated blast input obtained with
blastn -task megablast -query /home/aaron.dickey/data/contigs/skesa/mortest.fa -db nt -out 6test.txt -num_threads 12 -evalue 1e-20 -outfmt ‘6 std staxids scomnames sscinames sskingdoms’
and then:
/home/aaron.dickey/megan5/tools/blast2lca -f BlastTab -i 6test.txt -o all43_contig_assignments.txt